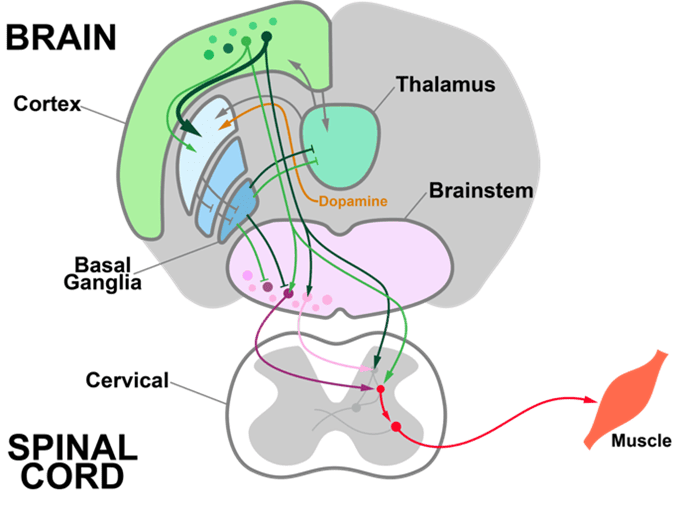
For decades, neurophysiology has emphasized the neural code, the information encoded in the firing of a given set of neurons. For example, the firing of a neuron might encode the face of a loved one or the movement of a muscle. But the neural code alone doesn’t tell us how the brain processes information. To understand how the brain recognizes a face or decides how to act, we need to know how neural circuits compute. Computations are the transformation of inputs into outputs.
Single-neuron Computations in Brain-wide Circuits (SCBC) is investigating neural computations by measuring patterns of input arriving at thousands of input synapses onto individual neurons, while also measuring their firing output. Analogous to how a computer algorithm is a series of input-output transformations, we aim to decompose brain computations into transformations of neural activity. Identifying these transformations requires measuring the inputs and the output of neurons simultaneously, a much harder technical challenge than identifying neural codes, for which only the output is needed.
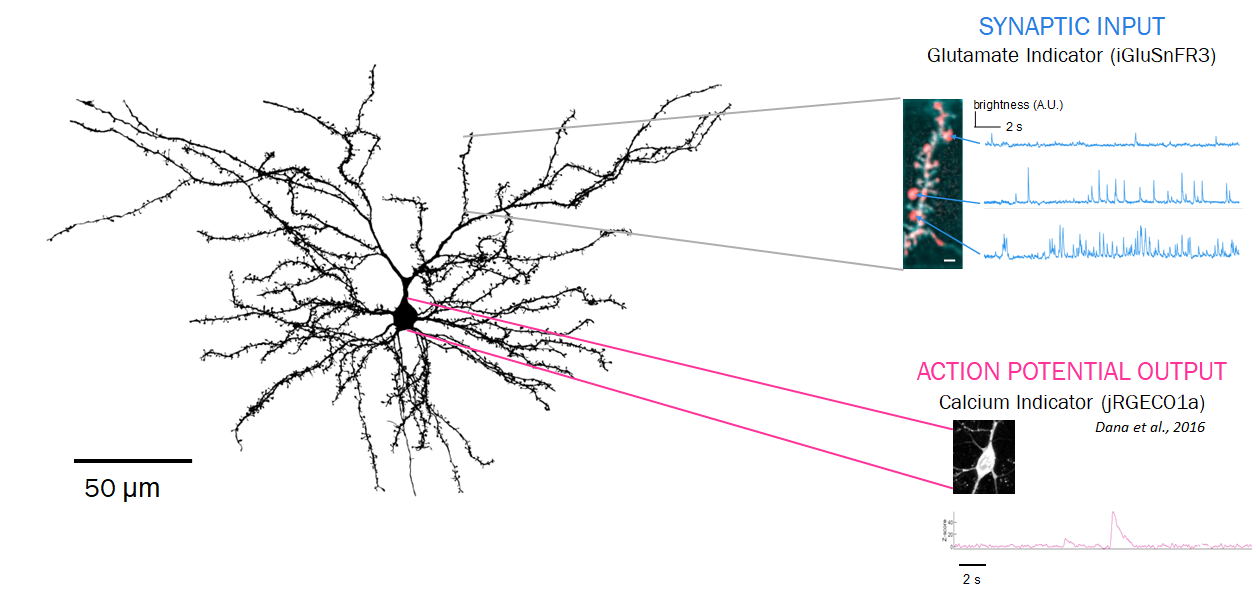
Neurons receive synaptic inputs at thousands of tiny spines along their 3-dimensional dendrites. Each synapse has a volume of less than 1 femtoliter. Specialized genetically encoded indicators and microscopes make it possible to measure these inputs, and the firing output they produce.
To make these unprecedented input-output measurements possible, we are developing new technologies that visualize synaptic activity at the microscopic scale in the living brain:
Neurotransmitter indicators
Neurons communicate at synapses by releasing brief (~1ms) bursts of neurotransmitter molecules, such as glutamate, GABA, dopamine, etc. To make these molecules visible, we’ve engineered genetically encoded indicators, proteins that fluoresce when they bind specific target molecules. These indicators are expressed on a neuron’s surface to visualize its synaptic inputs. In collaboration with other protein engineering groups, we are developing the next generation of indicators for glutamate, GABA, and several neuromodulators.
Our indicators are available from Addgene.
Microscopes for fast in vivo imaging
The living brain is opaque: it scatters and absorbs light. To image below its surface, we use two-photon microscopes based on scanning of pulsed infrared lasers. Recording synaptic input patterns is challenging for two-photon imaging because synaptic integration happens fast (>100 Hz) and synapses are distributed in 3 dimensions. Measuring activity from many synapses at speed requires fast and flexible ways to steer the microscope’s infrared laser. We have developed a microscope, SLAP2, based on a novel laser scanning system optimized for imaging large numbers of synapses, in the living, moving, brain.


Unlike conventional microscopes, SLAP2 can image any desired set of pixels in its field of view, in a time proportional to the number of pixels selected, without additional access time costs for each pixel (this makes SLAP2 different from microscopes based on acousto-optic scanners). SLAP2’s flexible scan system makes it 20-200 times faster for imaging synaptic activity than the best conventional microscopes and allows us to record from hundreds to thousands of synapses at the pace of neuronal computation. Importantly, SLAP2 has high-performance online motion correction to follow imaged synapses in 3D even as the brain moves during behavior.
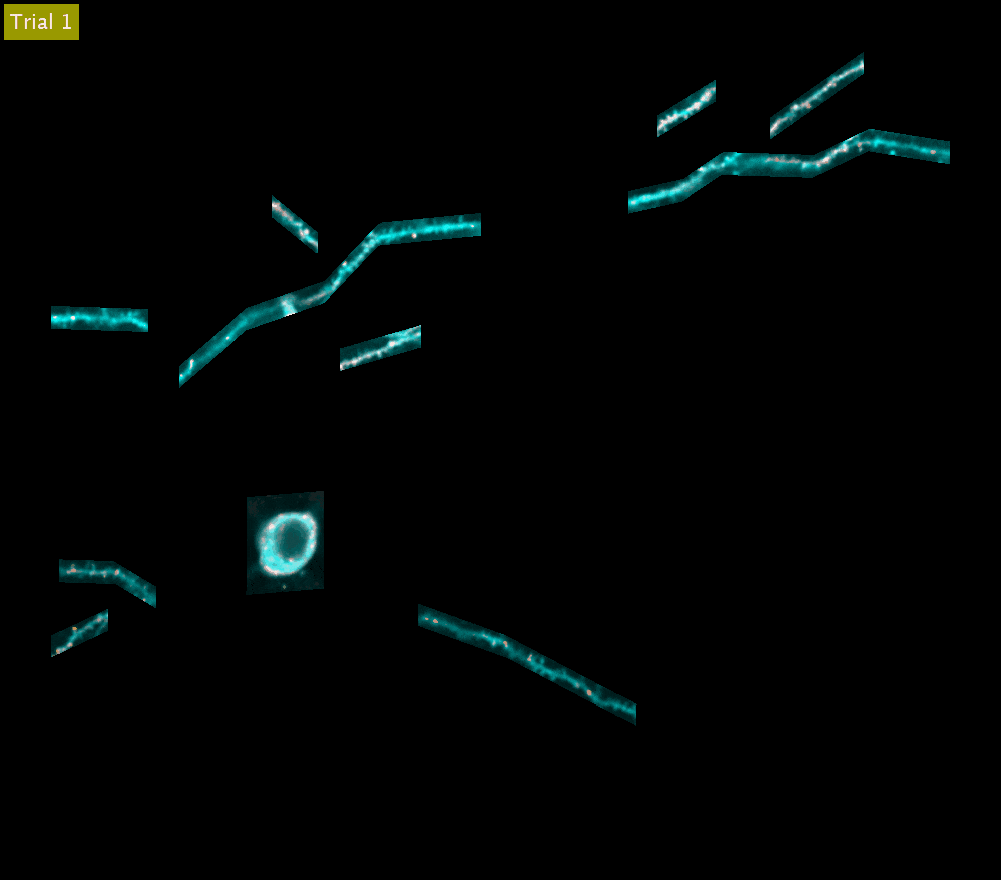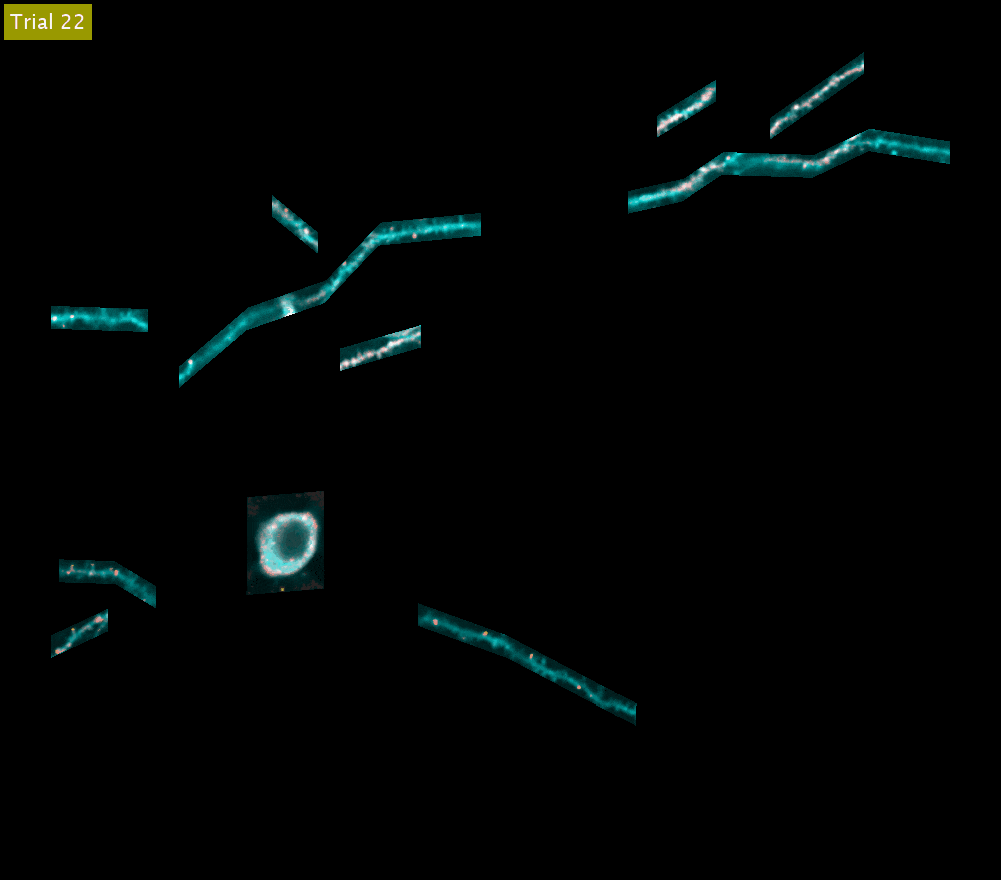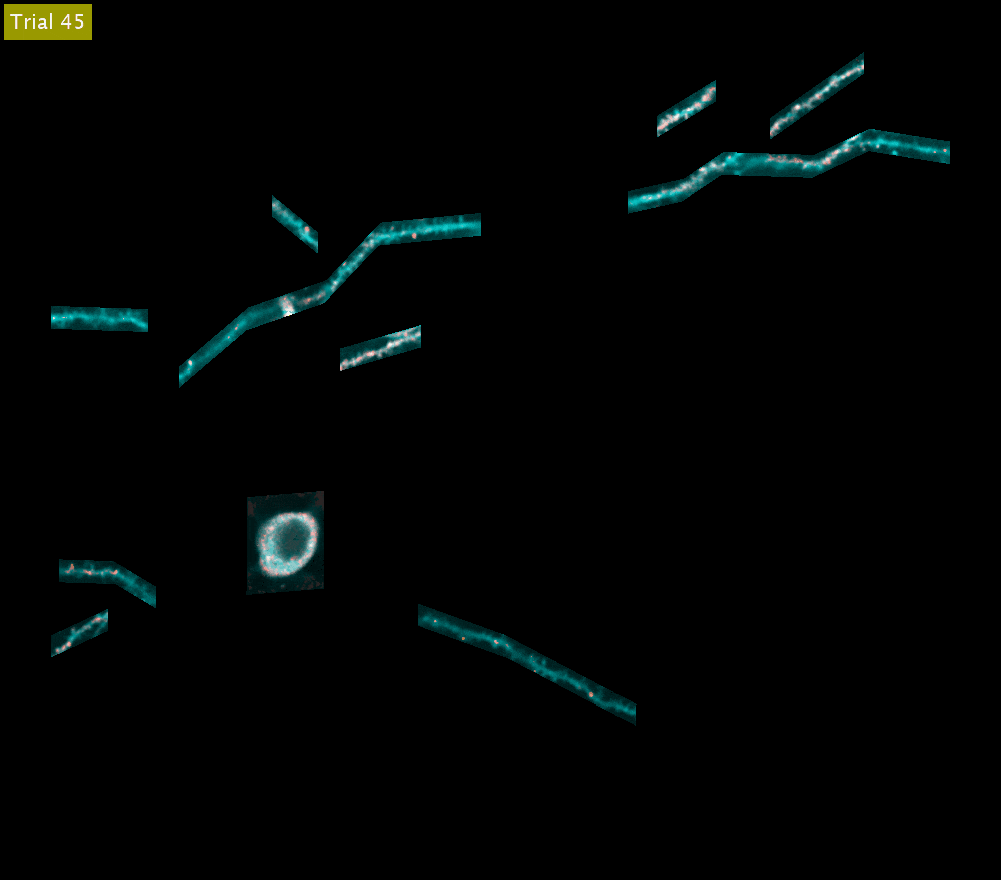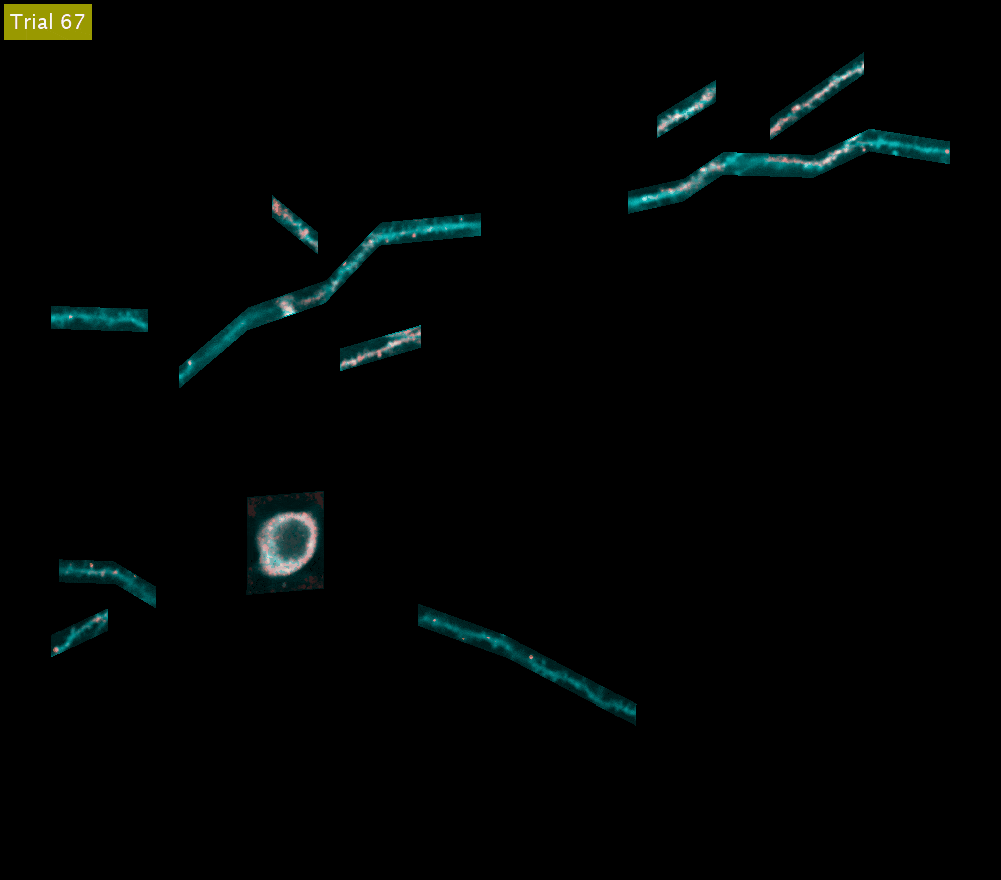
SLAP2 records from arbitrary sets of pixels across multiple sample planes. The video above shows iGluSnFR3 recordings (synaptic activity in red, structure in cyan) across behavioral trials from the cell body and dendrites of a Layer 2/3 neuron in the mouse motor cortex (collapsed into a single plane). These dendrites were imaged at 220 Hz.
SLAP2 microscope kits and software are now being distributed by MBF Biosciences.
OpenScope Steering Committee
The OpenScope Steering Committee convenes at least biannually to provide crucial direction for the OpenScope project, playing an essential role in ensuring that we effectively serve our broader community.