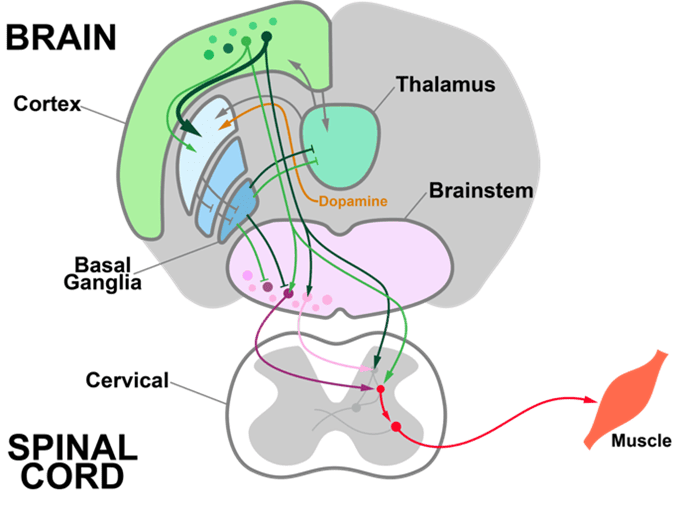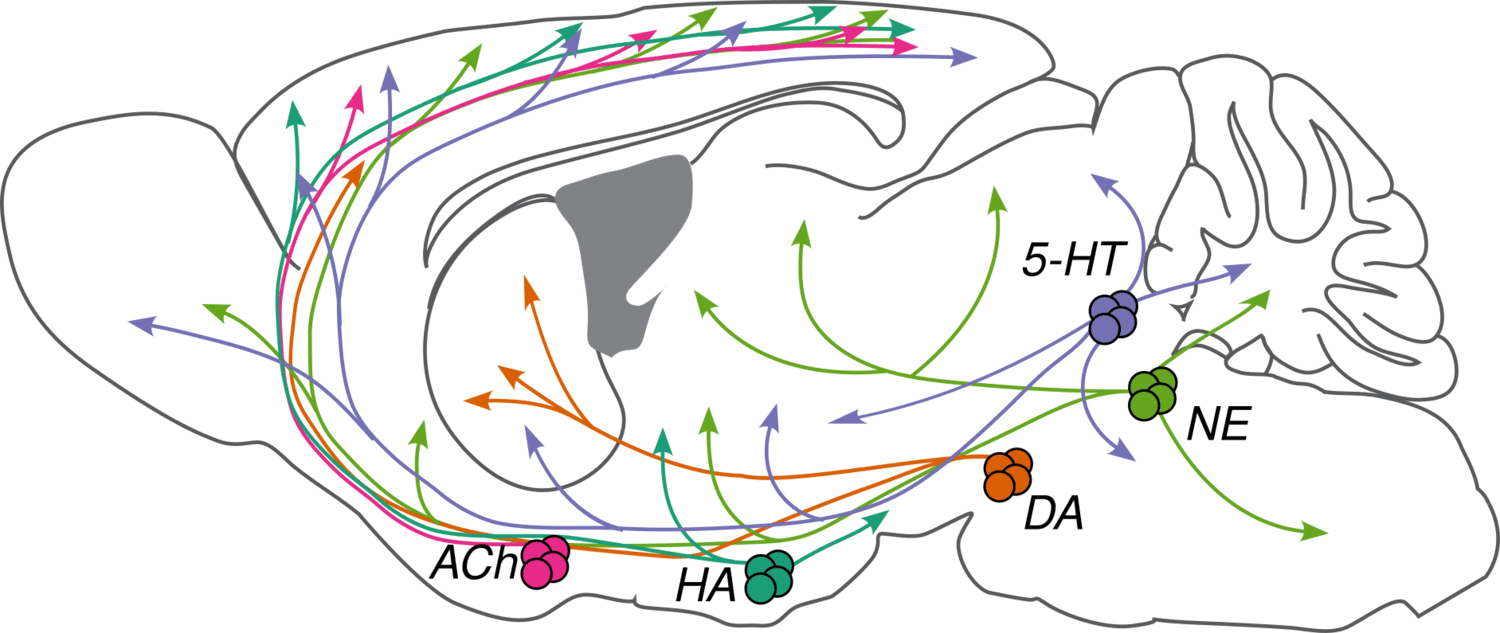
Neuromodulators (NMs) are neurotransmitters released by a small number of neurons (less than 0.001% of the neurons in our brain) with broad projections to most of the nervous system. They bind primarily to G-protein-coupled receptors on neurons and glia to activate intracellular signaling cascades and modulate the properties of membranes and synapses. The physiological processes triggered by NM release are often slow compared to fast synaptic transmission (hundreds of milliseconds or longer vs. less than ten milliseconds). These cells are largely conserved across vertebrates and have diverse functions in behavior, including modulating sleep-wake cycles, internal homeostasis, learning, arousal, and movement. Early theories that focused on specific behavioral conditions yielded to a more comprehensive view that NMs modulate all behavior, from gastric rhythms in invertebrates to mammalian cognition.
We are studying five major NMs: four biogenic amines—norepinephrine (NE), serotonin (5-HT), dopamine (DA), and histamine (HA)—and acetylcholine (ACh). They are primarily released by neurons in the locus coeruleus (NE), raphe nuclei (5-HT), ventral tegmental area and substantia nigra pars compacta (DA), tuberomammillary nucleus (HA), and several cholinergic areas across the brain, including the nucleus basalis (ACh). Many disorders, including depression, schizophrenia, drug addiction, and Parkinson's disease, appear to involve dysfunction of NM signaling.
We aim to address the following questions:
- What behavioral variables correlate with the firing of NM neurons and release across the brain?
- How do networks of neurons across the brain—with different NM receptors—use combinatorial NM input to drive learning and decision-making?
- What are the molecular and anatomical subclasses of NM neurons?
Answering these questions will require the following ingredients:
- Quantitative and controlled behaviors that engage NM neurons.
- Cell-type-specific electrophysiology from NM neurons.
- Calcium imaging from NM axons.
- Fluorescent sensors to measure NM release dynamics.
- Cell-type-specific manipulations of NM neurons.
- Reconstructions of NM neuron morphologies.
- Molecular-genetic tools to target subclasses of NM neurons.
What behavioral variables correlate with firing of NM neurons and release across the brain?
We are measuring NM neuron activity and NM release dynamics in multiple brain regions during foraging behaviors in mice. We are interested in behaviors that give us access to neural dynamics across brain regions, over multiple timescales. This requires a careful balance of well-controlled tasks that also tap into evolutionarily relevant behaviors across mammals, including mice. A core question is how the nervous system learns from recent experience in dynamic environments.

We focus on tasks that are amenable to quantitative modeling, including rich theories from reinforcement learning and foraging. One such task asks mice to freely choose among alternative actions that yield rewards with changing probabilities. These simple “dynamic foraging” tasks reveal how the nervous system uses action and reward history to choose future actions.
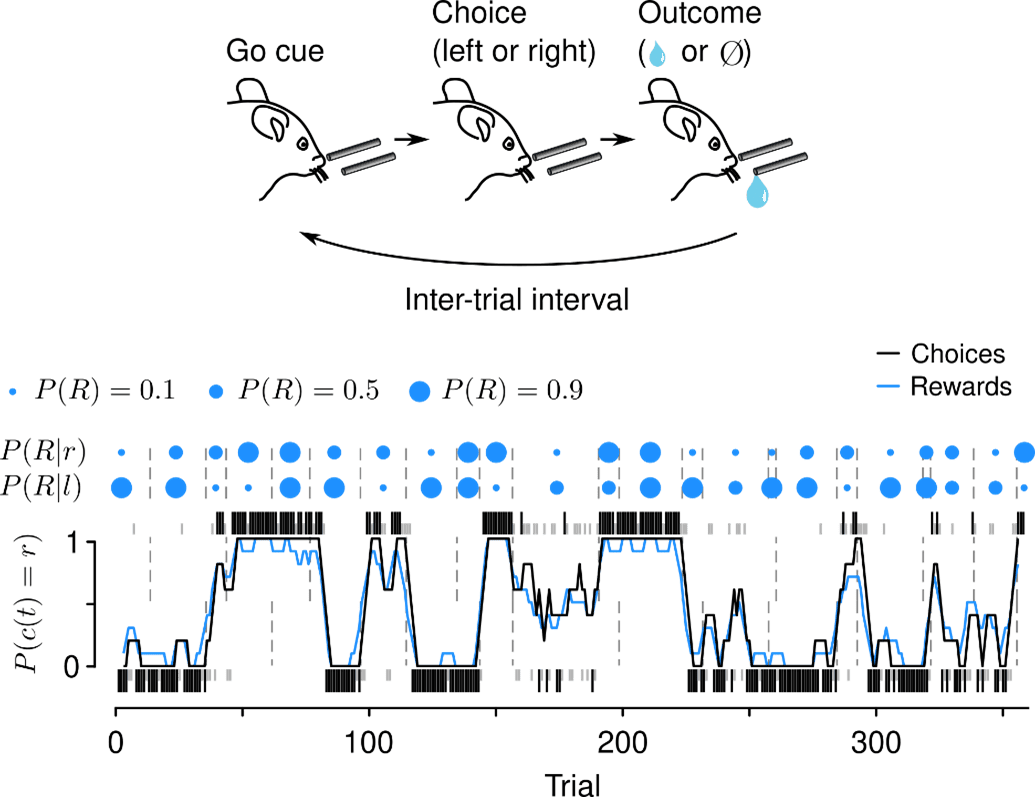
While mice forage for reward, we make two kinds of measurements. First, we record action potentials from LC-NE, DR-5-HT, VTA-DA, and SNc-DA neurons and study their correlations with behavior. We are interested in the diversity of these cells and their “receptive fields”: what behavioral events correlate with changes in firing rates of NE, 5-HT, and DA cells? We recently discovered that LC-NE neurons show two distinct patterns of activity during foraging.
%252520(1).png)
In parallel experiments, we are measuring the axonal activity of NM neurons across multiple target regions using calcium sensors and NM release across the brain using NM binding sensors.
How do networks of neurons across the brain—with different NM receptors—use combinatorial NM input to drive learning and decision-making?
Networks of neurons in the cerebral cortex, basal ganglia, and thalamus receive multiple NM inputs that adjust dynamics in these regions. How do NM inputs combine to change the activity of different cell types in these areas? We aim to record electrophysiological activity from multiple regions of the cortex, basal ganglia, and thalamus, while measuring and manipulating their NM inputs. These experiments will help us develop “input-output” functions for how NMs “tune the knobs” on brain-wide activity during behavior.
What are the molecular and anatomical subclasses of NM neurons?
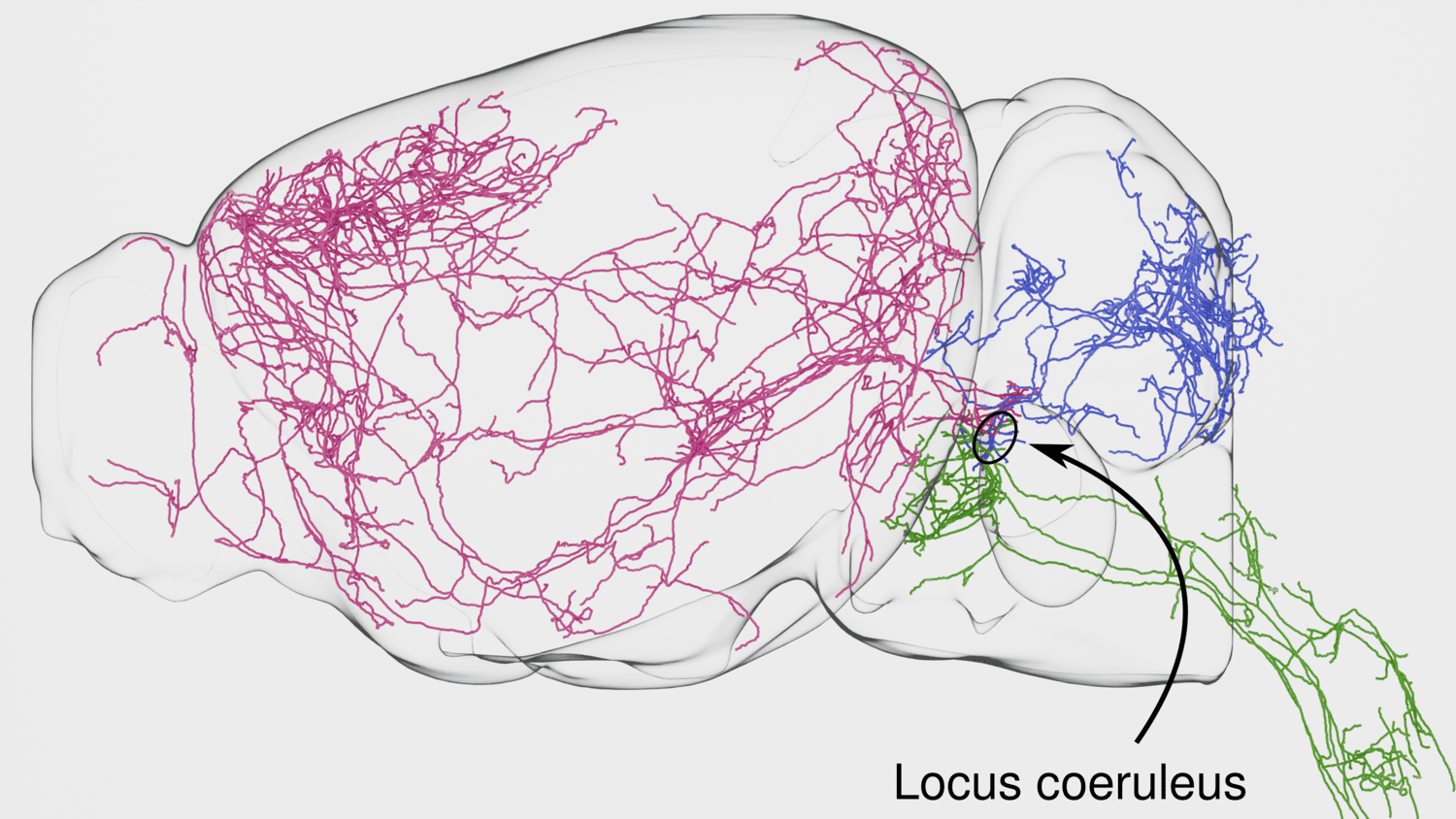
The classical view of NM neurons as monolithic projection systems has been superseded in recent years by the view that these systems are made up of subclasses of cells with distinct projection targets and molecular phenotypes. We are reconstructing the complete morphologies of NM cells—starting with LC-NE and DR-5-HT—to build a comprehensive map of their projections. We are correlating these morphologies with other molecular features of NM cells, using transcriptomics, multiomics, retrograde tracing, and other spatial sequencing techniques (BARseq, MAPseq).
OpenScope Steering Committee
The OpenScope Steering Committee convenes at least biannually to provide crucial direction for the OpenScope project, playing an essential role in ensuring that we effectively serve our broader community.